Phage (phage.mac)
Table of contents
Overview
The example simulates the irradiation of a DNA phage containing 141158 bp, placed in a cylinder with radius 3.5 um and height 7 um.
Geometry
Phage geometry is implemented in the provided macro file phage.mac. The file phage.txt describes the atom positions in the geometry.
Radical kill distance and direct interaction range are set to 4 nm and 4 angstrom, respectively.
# Geometry: size of World volume
/world/worldSize 9 um
# Geometry: creation
# - Side length for each placement
/dnageom/placementSize 50 50 50 nm
# - Scaling of XYZ in fractal definition file
/dnageom/fractalScaling 50 50 50 nm
# - Path to file that defines placement locations
/dnageom/definitionFile geometries/phage.txt
# - Set placement volumes
/dnageom/placementVolume turn geometries/1strand_50nm_turn.txt
/dnageom/placementVolume turntwist geometries/1strand_50nm_turn.txt true
/dnageom/placementVolume straight geometries/1strand_50nm_straight.txt
# Geometry: distance from base pairs at which radicals are killed
/dnageom/radicalKillDistance 4 nm
# Geometry: deposited energy accumulation range limit to start recording SBs from direct effects
/dnageom/interactionDirectRange 4.0 angstrom
The chromosome as region of interest for damage analysis is defined using:
/chromosome/add phage cyl 3500 7000 0 0 0 nm 0 0 0
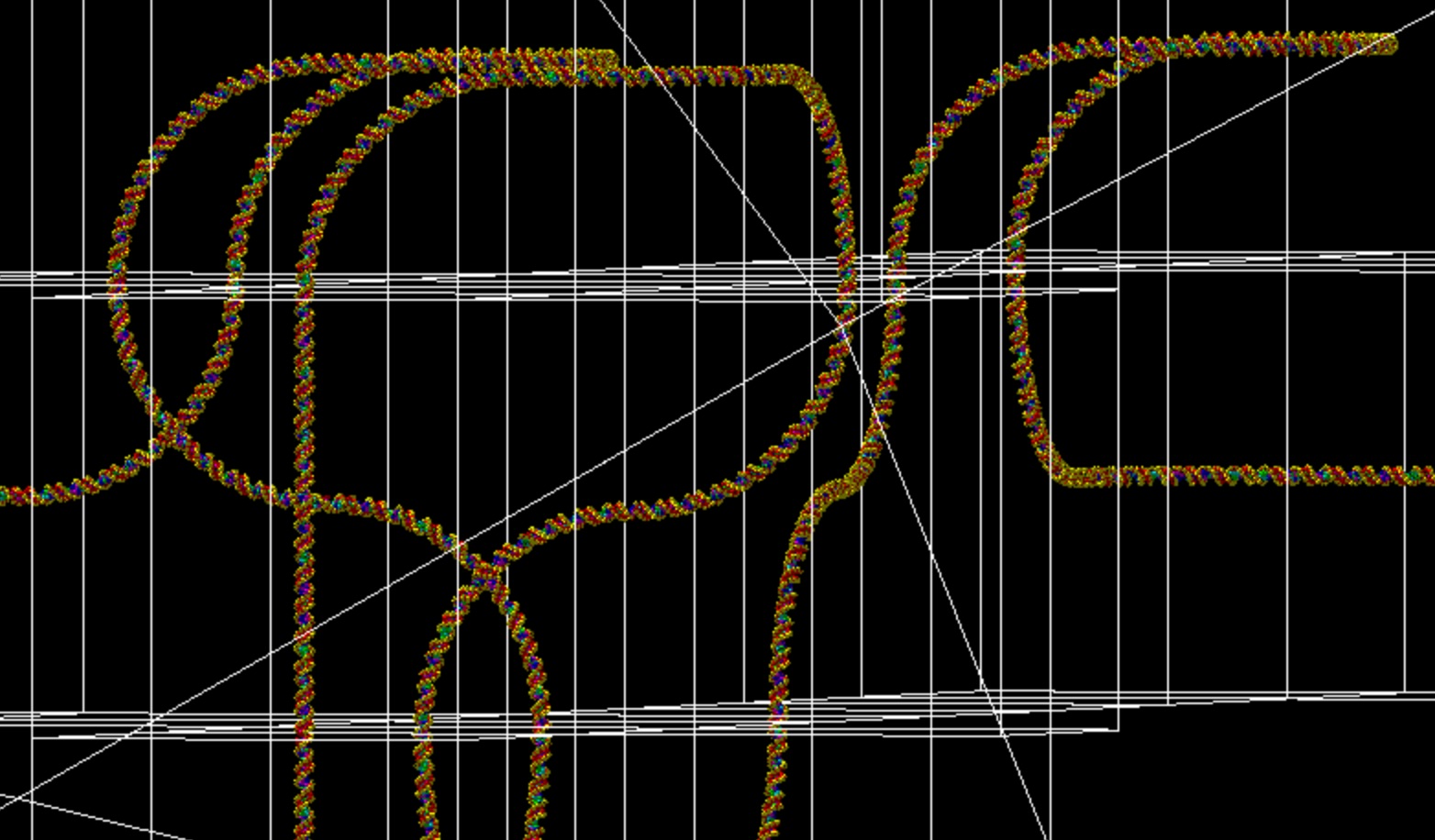
Example of phage geometry visualization.
Particle source
A proton plane circular source is used, shooting a parallel beam.
# Source geometry
/gps/pos/type Plane
/gps/pos/shape Circle
/gps/pos/centre 0 7000 0 nm
/gps/pos/rot1 0 0 1
/gps/pos/rot2 1 0 0
/gps/pos/radius 3500 nm
# Source particle, energy and angular distribution
/gps/particle proton
/gps/energy 2.5 MeV
/gps/direction 0 -1 0
# Beam on
/run/beamOn 10000
Damage model
The following settings are used:
/dnadamage/directDamageLower 5 eV
/dnadamage/directDamageUpper 37.5 eV
/dnadamage/indirectOHBaseChance 1.0
/dnadamage/indirectOHStrandChance 0.405
/dnadamage/inductionOHChance 0.00
/dnadamage/indirectHBaseChance 1.0
/dnadamage/indirectHStrandChance 0.0
/dnadamage/inductionHChance 0.00
/dnadamage/indirectEaqBaseChance 1.0
/dnadamage/indirectEaqStrandChance 0.0
/dnadamage/inductionEaqChance 0.00
Results
Output (see analysis) is analysed by using the phage.C ROOT macro file.
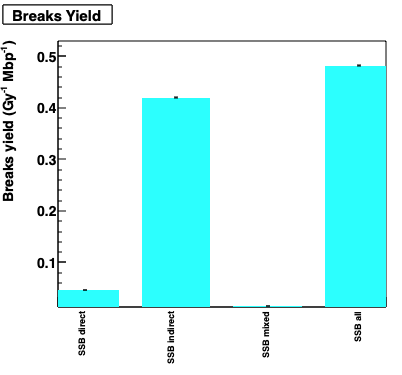
Example of damage simulation.
Reference
This geometry was developed during the ESA/BioRadIII project.