Repair model
This model calculates the accumulated proteins yield, by considering four principal DSB repair pathways.
These pathways are [1]:
- non-homologous end-joining (NHEJ),
- homologous recombination (HR),
- single-strand annealing (SSA), and
- alternative end-joining mechanism (Alt-NHEJ).
The number of non-repairable DSB/Gy/cell is needed (NcDSB), as well as the number of DSBs that is more probable to be repaired (NncDSB). More analytically:
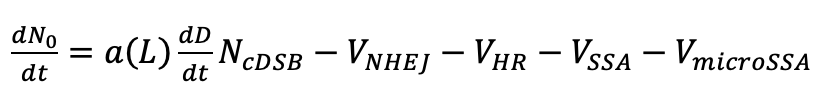
Where:
- N0 is the total number of DSB (NncDSB + NcDSB).
- VNHEJ, VHR, VSSA and VmicroSSA are mathematical variables that characterize the way that each model affects the way that DSBs are repaired.
- D is the dose (Gy).
- a(L) is the slope coefficient of linear dose dependence that describes DSB induction per unit of dose (/Gy/cell) and depends on LET.
User guide
To run the code, users need to open a terminal in the folder repair_survival_models containing the molecularDNArepair.py script
Then,
python3 molecularDNArepair.py
Input parameters
At line 13, users need to set the name of the output file, which is print in text format.
outputFile = "molecularDNArepair.txt"
At line 14, users need to set the name of the input file, which for the current version is in ROOT format. A future update of moleculardna example will include the option for using SDD files.
iRootFile = "../molecular-dna.root"
At line 18, users need to define the dimensions of the cell (x, y, z) in meters.
r3 = 7100*1e-09 * 2500*1e-09 * 7100*1e-09
At line 19, users need to define the length (in bp) of the DNA molecule model included in the simulation. The length of the DNA included in the “human cell” example is 6405886128 bp.
NBP = 6405886128
At line 20, the code calculates the mass of the cell used in the simulation. If another cell shape has been defined, other than the ellipsoid, the user needs to modify this calculation.
mass = 997 * 4 * 3.141592 * r3 / 3
Model parameters
At lines 106 - 173, the input parameters for the repair model have been included. The default values are those included in Belov et al. [2]. Users can modify these parameters. More extensive description for what each parameter stands for, can be found in the corresponding articles [1-3]. At lines 371 - 377, the values of additional data published in Chatzipapas et al. [3] have been included.
References
[1] Performance Evaluation for Repair of HSGc-C5 Carcinoma Cell Using Geant4-DNA, D. Sakata et al., Cancers 13 (2021) 6046 - link
[2] A quantitative model of the major pathways for radiation-induced DNA double-strand break repair, O. V. Belov et al., J. Theor. Biol., 366 (2015) 115-130 - link
[3] Simulation of DNA damage using Geant4-DNA: an overview of the “molecularDNA” example application, K. Chatzipapas et al., Prec. Radiat. Oncol. 7 (2023) 4-14 - link