Human cell (human_cell*.mac)
Table of contents
Overview
DNA damage induced by irradiation in a simplified human fibroblast cell can be simulated using the provided human_cell.mac macro file. A large amount of memory and computer performance will be required for this example. The parameters used in the macro file and shown in this page are further described in Phys. Med. 112 (2023) 102613 (link) and in Phys. Med. 127 (2024) 104389 (link).
Geometry
The default geometry (macro human_cell.mac) consists of a continuous chain defined by taking a basic Hilbert curve. This fractal is broken into cubic regions of straight and turned chromatin sections DNA placement. This chain is included in an ellipsoid with semi-dimensions 7.1 μm x 2.5 μm x 7.1 μm, which imitates a cell nucleus. Only cubes that are completely included in the ellipsoid are considered as parts of the chain, which length is 6.4 Gbp. Bp density of the produced cell corresponds to ~0.015 bp/nm3.
# Geometry: size of World volume
/world/worldSize 50 um
# Geometry: shape of the cell
/cell/radiusSize 14 2.5 14 um
# Chemistry: end time of chemistry stage
/scheduler/endTime 5.0 ns
# Chemistry: set maximum allowed zero time steps
/scheduler/maxNullTimeSteps 10000000
# Geometry: distance from base pairs at which radicals are killed
/dnageom/radicalKillDistance 9 nm
# Geometry: deposited energy accumulation range limit to start recording SBs from direct effects
/dnageom/interactionDirectRange 2.0 angstrom
# Geometry: creation
# - Side length for each placement
/dnageom/placementSize 75 75 75 nm
# - Scaling of XYZ in fractal definition file
/dnageom/fractalScaling 75 75 75 nm
# - Path to file that defines placement locations
/dnageom/definitionFile geometries/cube-centred-X-8.txt
# - Set placement volumes
/dnageom/placementVolume turn geometries/turned_solenoid_750_withHistone.txt
/dnageom/placementVolume turntwist geometries/turned_twisted_solenoid_750_withHistone.txt true
/dnageom/placementVolume straight geometries/straight_solenoid_750_withHistone.txt
The DNA geometrical model and the irradiation setup in the simulation are as follows:
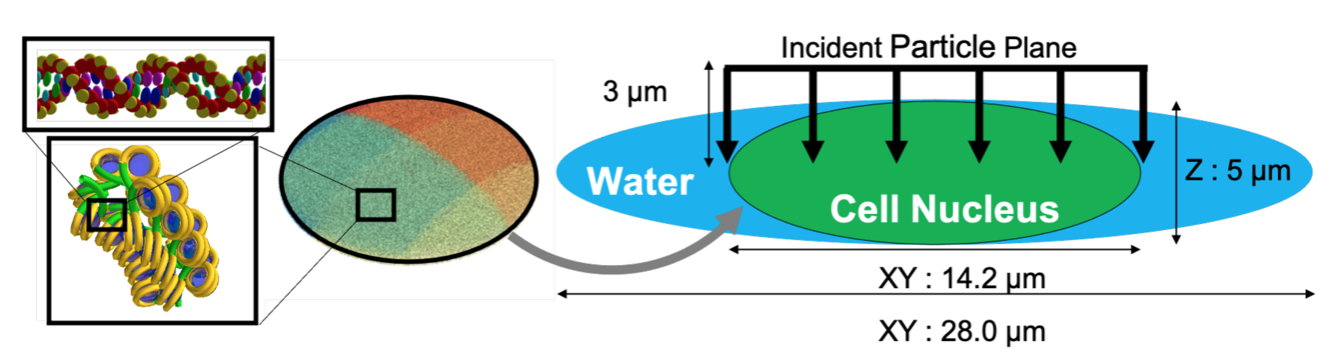
Left: The 3D geometry of the cell nucleus (14.2 μm x 14.2 μm x 5 μm) used in the human_cell.mac macro file, showing the continuous fractal interior. Right: The beam geometry used in the simulation, showing the incident protons as a parallel beam.
The chromosome as region of interest for damage analysis is defined using:
/chromosome/add cell ellipse 7100 2500 7100 0 0 0 nm 0 0 0
Particle source
A proton source plane with circle radius 7.1 um is located 3 μm from the cell center.
# Source geometry
/gps/pos/type Plane
/gps/pos/shape Circle
/gps/pos/centre 0 3000 0 nm
/gps/pos/rot1 0 0 1
/gps/pos/rot2 1 0 0
/gps/pos/radius 7100 nm
/gps/direction 0 -1 0
# Source particle
/gps/particle proton
# Source energy
/gps/energy 0.4 MeV
# Beam on
/run/beamOn 215
Damage model
Direct damage model sets 5 eV for the lower break threshold and 37.5 eV for the upper break threshold. A probability of 40.5% is set for the production of strand break.
/dnadamage/directDamageLower 5 eV
/dnadamage/directDamageUpper 37.5 eV
/dnadamage/indirectOHBaseChance 1.0
/dnadamage/indirectOHStrandChance 0.405
/dnadamage/inductionOHChance 0.0
/dnadamage/indirectHBaseChance 1.0
/dnadamage/indirectHStrandChance 0.0
/dnadamage/inductionHChance 0.0
/dnadamage/indirectEaqBaseChance 1.0
/dnadamage/indirectEaqStrandChance 0.0
/dnadamage/inductionEaqChance 0.0
Results
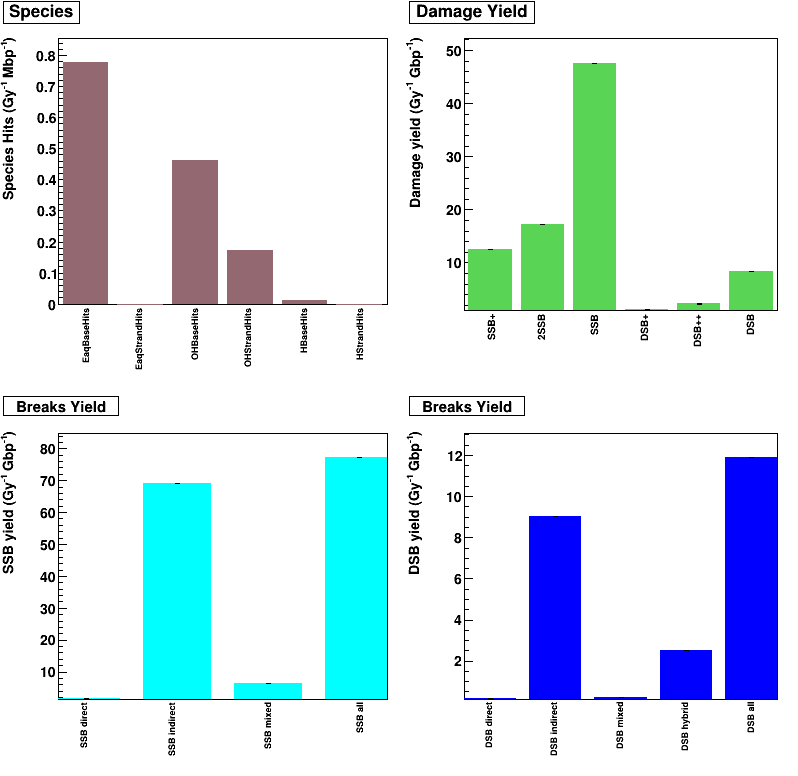
Output (see analysis) is analysed by using the human_cell.C ROOT macro file.
-
Species Hits (/Gy/Mbp) is defined by radical + DNA reactions, for example: EaqStrandHits is e_aq + DNA backbone
-
Damage yield (/Gy/Gbp) is defined by DNA damage complexity (see classification model)
-
Breaks yield (/Gy/Gbp) is showed for each break type (direct SSB, indirect SSB, DSB,…).
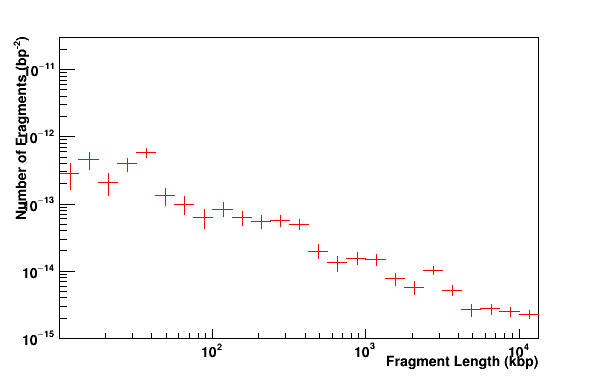
Fragments distribution of DNA. A fragment is defined by a distance between two DSBs.
List of Geant4 macros
The example provides four macros to simulate the cellular irradiation of three different cell types, see Phys. Med. 112 (2023) 102613 (link) and Phys. Med. 127 (2024) 104389 (link) :
-
human_cell.mac : this is the default geometry of a simplified human fibroblast, as described above (ellipsoid with semi-dimensions 7.1 μm x 2.5 μm x 7.1 μm)
-
human_cell_HTB177.mac : this is a simplified model of a human large cell lung carcinoma, represented by an ellipsoid with semi-dimensions 8.55 μm x 2.5 μm x 6.425 μm
-
human_cell_MCF7.mac : this is a simplified model of a human breast adenocarcinoma, represented by an ellipsoid with semi-dimensions 7.005 μm × 2.50 μm × 5.30 μm
-
human_cell_chromosomes.mac : this is a simplified model of a human human fibroblast containing individual chromosomes, represented by an ellipsoid with semi-dimensions 10.575 μm × 3.45 μm × 10.575 μm
List of ROOT macros
Three ROOT macros are provided to analyze the simulation results obtained with the above Geant4 macros : human_cell.C, human_cell_alphas.C and human_cell_chromosomes.C.