Bacterial cell (ecoli.mac)
Table of contents
Overview
This example uses the E. coli bacterium geometry, which imitates the genome of the bacterium. The simulation can be run using the ecoli.mac macro file.
Geometry
The genome has been produced using four side-by-side Hilbert curve fractals (see FractalDNA). This creates 16,384 placement volumes that are assumed to be cubic boxes with a side length of 50 nm. This was composed of 7,200 straight segments, and 5,652 turned segments of DNA. We only placed placement volumes that fell inside an ellipsoid with a semi-major axis of 900 nm and two equal semi-minor axes of 400 nm, creating an elliptical geometry that corresponded roughly to the dimensions of an E. coli bacterium containing 4,864 placement volumes. The final geometry (Figure) contained 4.63 Mbp, similar again to the length of an E. coli genome. Here’s an example of a macro file:
# Geometry: size of World volume
/world/worldSize 8 um
# Chemistry: end time of chemistry stage
/scheduler/endTime 1 us
# Geometry: optimisation of voxelisation
/dnageom/setSmartVoxels 1
# Geometry: distance from base pairs at which radicals are killed
/dnageom/radicalKillDistance 4 nm
# Geometry: deposited energy accumulation range limit to start recording SBs from direct effects
/dnageom/interactionDirectRange 6 angstrom
# Geometry: creation
# - Side length for each placement
/dnageom/placementSize 50 50 50 nm
# - Scaling of XYZ in fractal definition file
/dnageom/fractalScaling 50 50 50 nm
# - Path to file that defines placement locations
/dnageom/definitionFile geometries/bacteria-XFXFXFX-4.txt
# - Set placement volumes
/dnageom/placementVolume turn geometries/8strands_50nm_turn.txt
/dnageom/placementVolume turntwist geometries/8strands_50nm_turn.txt true
/dnageom/placementVolume straight geometries/8strands_50nm_straight.txt
# Analysis: add ellipsoid chromosomal region of interest, with the name "bacteria"
/chromosome/add bacteria ellipse 900 400 400 0 0 0 nm 0 0 0
Particle source
Electrons are simulated coming from an ellipse enclosing the bacterial cell (of the same dimensions as the cell) with energy 9.999 keV. The angular distribution of electron trajectories coming from the cell surface follows a cosine law, which simulates an isotropic radiation environment.
# Source geometry
/gps/pos/type Surface
/gps/pos/shape Ellipsoid
/gps/pos/centre 0 0 0 nm
/gps/pos/halfx 900 nm
/gps/pos/halfy 400 nm
/gps/pos/halfz 400 nm
/gps/ang/type cos
# Source particle and energy
/gps/particle e-
/gps/energy 9.999 keV
# Beam on
/run/beamOn 50000
Damage model
Direct damage model uses the 17.5 eV for lower and upper break threshold. The probability of 40 % for the production of strand break by OH (OH + 2-deoxyribose) is applied.
/dnadamage/directDamageLower 17.5 eV
/dnadamage/directDamageUpper 17.5 eV
/dnadamage/indirectOHBaseChance 1.0
/dnadamage/indirectOHStrandChance 0.4
/dnadamage/inductionOHChance 0.0
/dnadamage/indirectHBaseChance 1.0
/dnadamage/indirectHStrandChance 0.4
/dnadamage/inductionHChance 0.0
/dnadamage/indirectEaqBaseChance 1.0
/dnadamage/indirectEaqStrandChance 0.4
/dnadamage/inductionEaqChance 0.0
Results
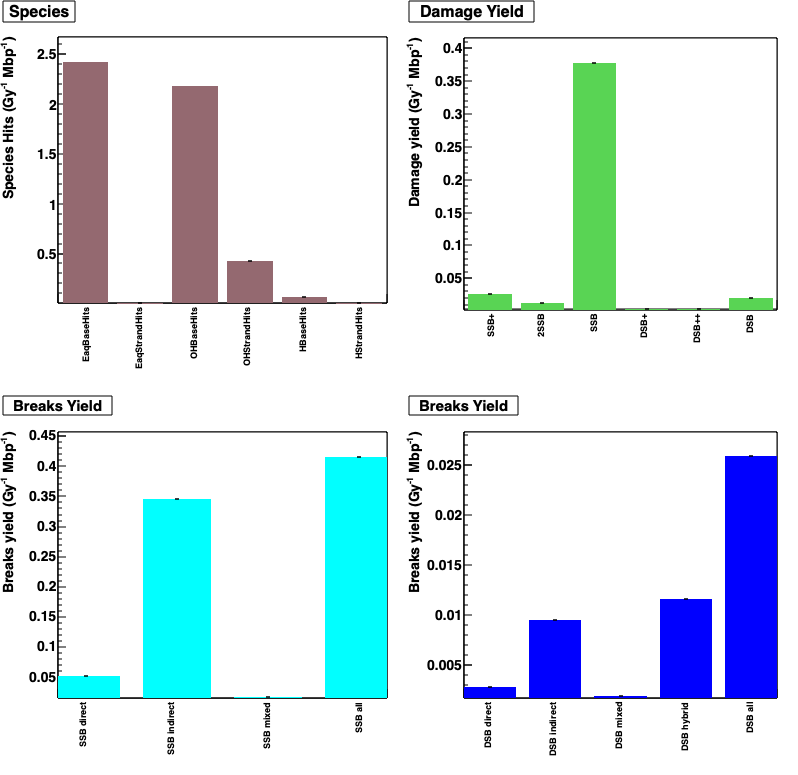
Output (see analysis) is analysed by using ecoli.C ROOT macro file.
-
Species hits (/Gy/Mbp) is defined by radical + DNA reactions, for example: EaqStrandHits is e_aq + DNA backbone
-
Damage yield (/Gy/Mbp) is defined by DNA damage complexity (see classification model)
-
Breaks yield (/Gy/Mbp) is showed for each break type (direct SSB, indirect SSB, DSB,…).
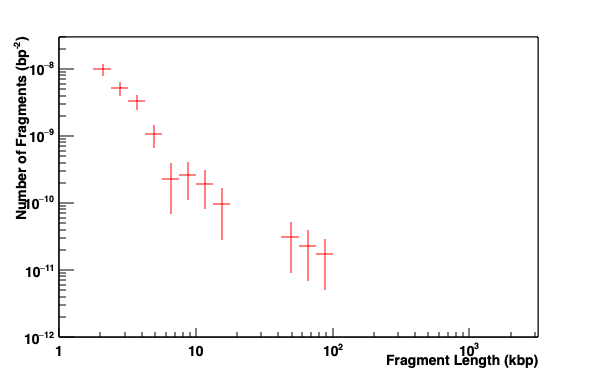
Fragments distribution of DNA. A fragment is defined by a distance between two DSBs.
Important notes
- The physics and chemistry models used since the 2018 publication of this model have evolved significantly, making comparison to past works difficult. Optimization of simulation parameters is thus required.
- Further, an issue was identified in the geometry implementation of the 2018 work that is addressed in this corrigendum.