DNA placements
The DNA placements here were built using the FractalDNA package in Python. Accompanying each geometry is the Python code to generate it.
Single DNA segments
Straight and turned segments for a 50nm box.
Generating code
from fractaldna.dna_models import dnachain
import numpy as np
bp_separation = dnachain.BP_SEPARATION # 3.32Å
side_length_nm = 50 # nm
num_basepairs_straight = int( side_length_nm / (0.1*bp_separation) )
num_basepairs_turned = int((side_length_nm * np.pi / 4.) / (0.1*bp_separation))
chain_straight = dnachain.DNAChain(
''.join(np.random.choice(['G', 'A', 'T', 'C'], num_basepairs_straight))
)
chain_turned = dnachain.TurnedDNAChain(
''.join(np.random.choice(['G', 'A', 'T', 'C'], num_basepairs_turned))
)
chain_turned_twisted = dnachain.TurnedTwistedDNAChain(
''.join(np.random.choice(['G', 'A', 'T', 'C'], num_basepairs_turned))
)
chain_straight.to_frame().to_csv('50nm_straight.csv', sep=' ', index=False)
chain_turned.to_frame().to_csv('50nm_turn.csv', sep=' ', index=False)
chain_turned_twisted.to_frame().to_csv('50nm_turn_twist.csv', sep=' ', index=False)
Simple DNA segments with multiple strands
Sometimes to make denser DNA, you will want to increase the amount of DNA in a voxel.
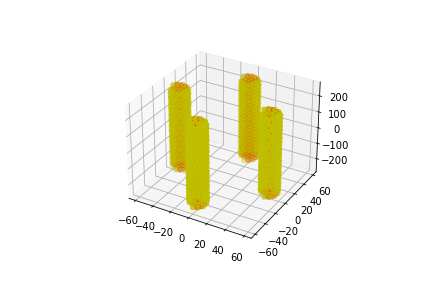
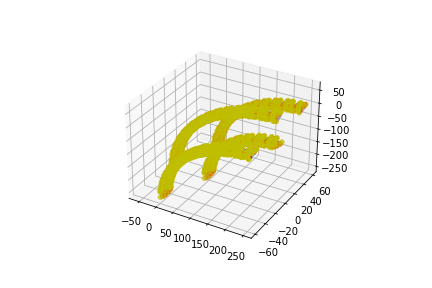
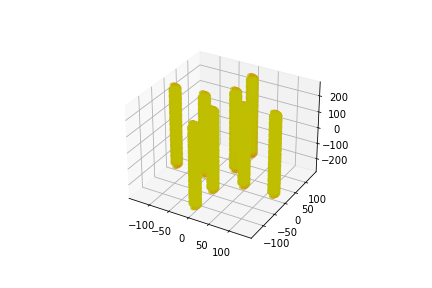
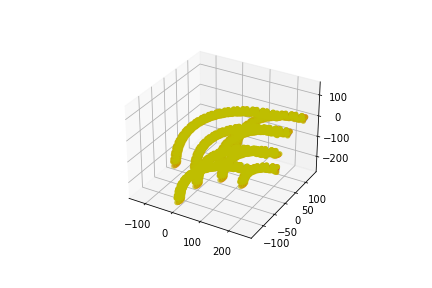
The following placements contain 4 parallel DNA strands.
Generating code
bp_separation = dnachain.BP_SEPARATION # 3.32Å
side_length_nm = 50 # nm
num_basepairs_straight = int( side_length_nm / (0.1*bp_separation) )
num_basepairs_turned = int((side_length_nm * np.pi / 4.) / (0.1*bp_separation))
strand_separation = 100 # angstroms
chain4_straight = dnachain.FourStrandDNAChain(
''.join(np.random.choice(['G', 'A', 'T', 'C'], num_basepairs_straight)),
strand_separation
)
chain4_turned = dnachain.FourStrandTurnedDNAChain(
''.join(np.random.choice(['G', 'A', 'T', 'C'], num_basepairs_turned)),
strand_separation
)
chain4_turned_twisted = dnachain.FourStrandTurnedDNAChain(
''.join(np.random.choice(['G', 'A', 'T', 'C'], num_basepairs_turned)),
strand_separation,
twist=True
)
chain4_straight.to_frame().to_csv('results/50nm_4_straight.csv', sep=' ', index=False)
chain4_turned.to_frame().to_csv('results/50nm_4_turn.csv', sep=' ', index=False)
chain4_turned_twisted.to_frame().to_csv('results/50nm_4_turn_twist.csv', sep=' ', index=False)
The following placements contain 8 parallel DNA strands.
Generating code
%%capture
bp_separation = dnachain.BP_SEPARATION # 3.32Å
side_length_nm = 50 # nm
num_basepairs_straight = int(side_length_nm / (0.1 * bp_separation))
num_basepairs_turned = int((side_length_nm * np.pi / 4.0) / (0.1 * bp_separation))
strand_separation_1 = 100 # angstroms
strand_separation_2 = 250 # angstroms
chain8_straight = dnachain.EightStrandDNAChain(
"".join(np.random.choice(["G", "A", "T", "C"], num_basepairs_straight)),
strand_separation_1,
strand_separation_2,
turn=False,
twist=False
)
chain8_turned = dnachain.EightStrandDNAChain(
"".join(np.random.choice(["G", "A", "T", "C"], num_basepairs_turned)),
strand_separation_1,
strand_separation_2,
turn=True,
twist=False
)
chain8_turned_twisted = dnachain.EightStrandDNAChain(
"".join(np.random.choice(["G", "A", "T", "C"], num_basepairs_turned)),
strand_separation_1,
strand_separation_2,
turn=True,
twist=True
)
chain8_straight.to_frame().to_csv("results/50nm_8_straight.csv", sep=" ", index=False)
chain8_turned.to_frame().to_csv("results/50nm_8_turn.csv", sep=" ", index=False)
chain8_turned_twisted.to_frame().to_csv(
"results/50nm_8_turn_twist.csv", sep=" ", index=False
)
Solenoidal DNA segments
You can also make solenoidal DNA.
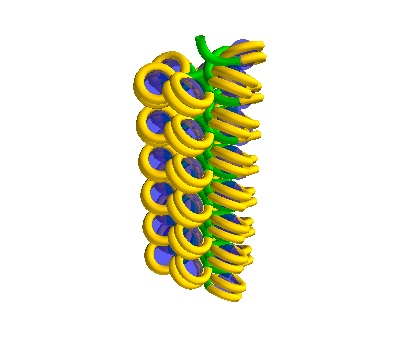
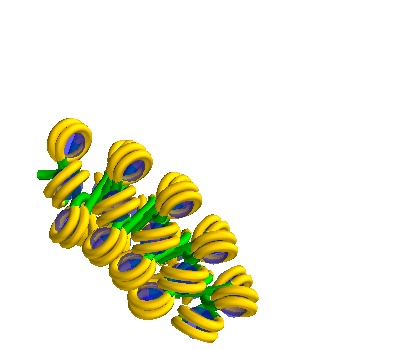
The following placements contain Solenoidal DNA
Generating code
from fractaldna.dna_models import dnachain
side_length = 750 # angstrom
radius_solenoid = 100 # angstrom
nhistones = 38 # histones
solenoid_straight = dnachain.Solenoid(
voxelheight=side_length, radius=radius_solenoid, nhistones=nhistones
)
solenoid_turned = dnachain.TurnedSolenoid(
voxelheight=side_length, radius=radius_solenoid, nhistones=nhistones
)
solenoid_turned_twisted = dnachain.TurnedSolenoid(
voxelheight=side_length, radius=radius_solenoid, nhistones=nhistones, twist=True
)
# centre around (x,y,z)=(0,0,0)
solenoid_straight.translate([0, 0, -side_length/2.])
solenoid_turned.translate([0, 0, -side_length/2.])
solenoid_turned_twisted.translate([0, 0, -side_length/2.])
solenoid_straight.to_frame().to_csv('results/solenoid_straight.csv', sep=' ', index=False)
solenoid_turned.to_frame().to_csv('results/solenoid_turned.csv', sep=' ', index=False)
solenoid_turned_twisted.to_frame().to_csv('results/solenoid_turned_twisted.csv', sep=' ', index=False)
Multiple solenoidal volumes
Here we make volumes that contain 4 solenoids:
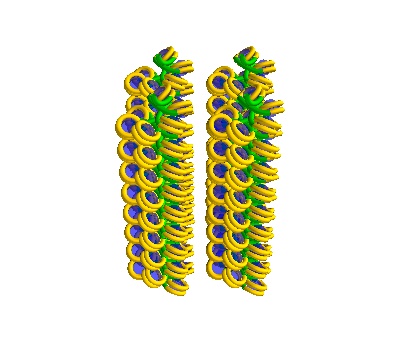
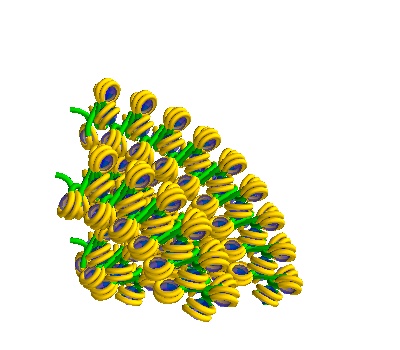
The following placements contain solenoidal DNA
Generating code
side_length = 1000 # angstrom
radius_solenoid = 100 # angstrom
nhistones = 51 # histones
separation = 250 # angstroms
solenoid4_straight = dnachain.MultiSolenoidVolume(
voxelheight=side_length,
separation=separation,
radius=radius_solenoid,
nhistones=nhistones,
chains=[1, 2, 3, 4],
turn=False,
twist=False,
)
solenoid4_turned = dnachain.MultiSolenoidVolume(
voxelheight=side_length,
separation=separation,
radius=radius_solenoid,
nhistones=nhistones,
chains=[1, 2, 3, 4],
turn=True,
twist=False
)
solenoid4_turned_twisted = dnachain.MultiSolenoidVolume(
voxelheight=side_length,
separation=separation,
radius=radius_solenoid,
nhistones=nhistones,
chains=[1, 2, 3, 4],
turn=True,
twist=True
)
# centre around (x,y,z)=(0,0,0)
solenoid4_straight.translate([0, 0, -side_length/2.])
solenoid4_turned.translate([0, 0, -side_length/2.])
solenoid4_turned_twisted.translate([0, 0, -side_length/2.])
solenoid4_straight.to_frame().to_csv('results/solenoid4_straight.csv', sep=' ', index=False)
solenoid4_turned.to_frame().to_csv('results/solenoid4_turned.csv', sep=' ', index=False)
solenoid4_turned_twisted.to_frame().to_csv('results/solenoid4_turned_twisted.csv', sep=' ', index=False)