Table of contents
Pre-chemical stage
The energy transfer induced by ionizing radiation in a water medium occurs rapidly (on a scale of femtoseconds (fs)) during the physical stage of water radiolysis and is followed by the formation of radiolysis species. These species are created in a very short time (from femtoseconds (fs) to picoseconds (ps)), mainly through electronic events during the pre-chemical stage. These events—such as thermalization, solvation of sub-excitation electrons, electronic hole migration, and fast electronic recombination— can lead to chemical bond breaks and produce species.
The excited H2O* and ionized H2O+ water molecules are dissociated into radical species based on dissociation channels used in Geant4-DNA [1][2][3].
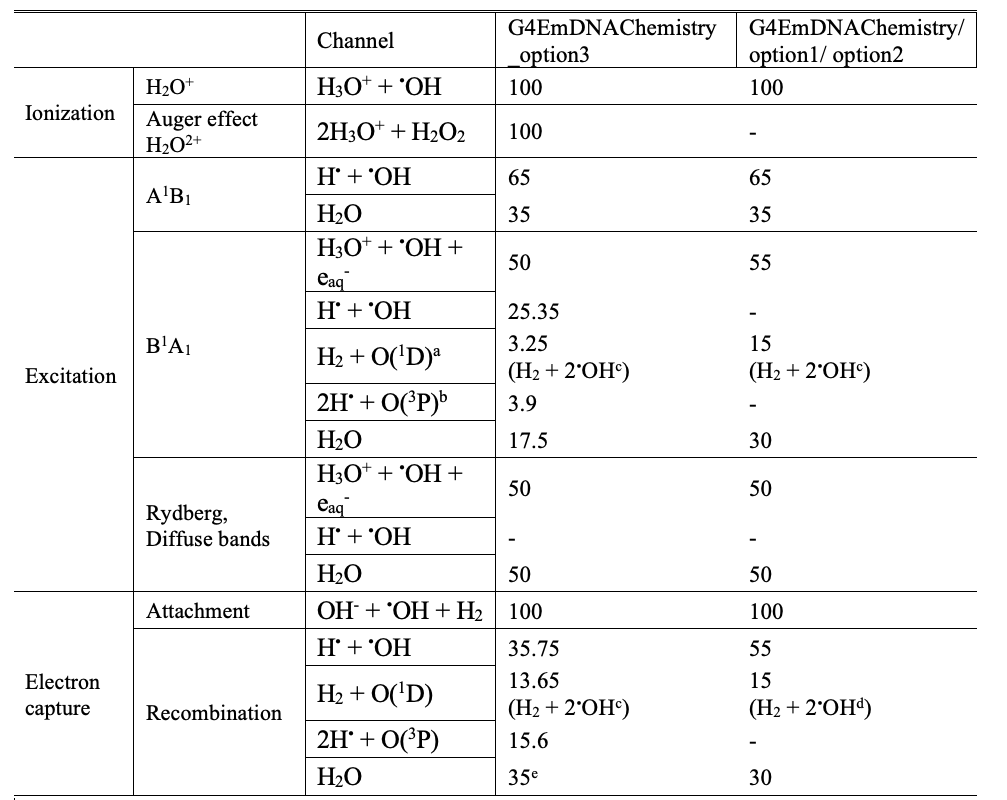
The molecularDNA example is using the “option3” dissociation channels as default.
Time step models
Chemistry stage starts from 1 ps for all radio-induced reactive species which are assumed to be hard spherical particles while water is considered as a continuum. In this stage, time step models describe an action (reaction or diffusion) of species during a discretized time step. In the molecularDNA example, the time step models determinate a competition between free radical species recombination and indirect DNA damages.
Synchronous IRT method
The IRT method is based on the “Independent Pair Approximation”; thus, reactive pairs are assumed independent, that is, the reaction time between any reactant pairs does not depend on the other reactants present in the medium. Under this assumption, the reaction time is sampled from the reaction probability distributions of the reactant pairs that mainly depend on initial pair distance. The IRT method determines the minimum time to the next reaction. Reactive products created by reactions that have occurred can undergo reactions with other reactants. These new reactions then need to be considered and included in the possible reactions.
While this is a considerable advantage in terms of computing time, the spatial–temporal information of the system is not simulated explicitly. As a complementary extension, synchronous IRT (or IRT-syn) implementation calculates a time step using IRT method for the next reaction that should occur. The reactive products created in this reaction and the remaining molecules are considered explicitly together to diffuse for the time step. Then, based on their new positions, the new random reaction times are re-evaluated sequentially for all the radicals in the system and the new minimum reaction time and corresponding reaction is selected for next time step. This procedure is repeated until the end time of simulation.
By default, a maximum time step of 500 ps is applied to all chemistry simulations [4], in the TimeStepAction class. This means that the chemical reactions expected to occur beyond 500 ps are ignored, all remainig molecules undergo a 500 ps diffusion, and the table of reactions is fully recalculated.
Reaction rates between free radicals and DNA
Indirect damage occurs from the chemical reaction between a radical and a DNA molecule (see the table below). To induce indirect strand breaks, the chemical reaction occurs between the •OH radical and the 2-deoxyribose-phosphate group. The probabilities to induce a single strand break are described in the Indirect Damage through DamageModel class.
| Reaction | Reaction rate (109 M-1s-1)[5] |
|---|---|
| 2-deoxyribose + •OH | 1.8 |
| Adenine + •OH | 6.1 |
| Guanine + •OH | 9.2 |
| Thymine + •OH | 6.4 |
| Cytosine + •OH | 6.1 |
| 2-deoxyribose + e-aq | 0.01 |
| Adenine + e-aq | 9.0 |
| Guanine + e-aq | 14.0 |
| Thymine + e-aq | 18.0 |
| Cytosine + e-aq | 13.0 |
| 2-deoxyribose + H• | 0.029 |
| Adenine + H• | 0.10 |
| Thymine + H• | 0.57 |
| Cytosine + H• | 0.092 |
Use of parallel worlds
The Geant4 chemistry module has difficulty dealing with complicated geometries due to dissociation processes, which can place the products of the molecular dissociation of an energetic molecule way from the dissociating molecule and geometry navigation, which requests computation time. To avoid having too many geometrical boundaries in chemistry simulations, all the physical volumes are placed in a separate parallel world, using the layered geometries offered by Geant4 (Enger et al., 2012). Thus, the physically placed DNA molecules described in this section are only seen by physical processes, and their boundaries are effectively ignored by chemistry. At each time step, chemical species are requests to look up nearby DNA molecules using an octree data structure.
Tips
Activate chemistry module using :
/chem/activate true # false for deactivation
The chemistry stage is simulated until 1 microsecond (by default). Users can decide the end time by using :
/scheduler/endTime 4 ns # set 4 nanosecond at which the simulation stops
To print reactions
/scheduler/verbose 1
References
[1] Modeling radiation chemistry in the Geant4 toolkit, M. Karamitros et al., Prog. Nucl. Sci. Tec. 2 (2011) 503-508 - link
[2] Geant4-DNA simulation of the pre-chemical stage of water radiolysis and its impact on initial radiochemical yields, W.-G. Shin et al., Phys. Med. 88, 2021, 86-90 - link
[3] Review of chemical models and applications in Geant4-DNA: Report from the ESA BioRad III Project, H. N. Tran et al., Med. Phys. (2024) in press - link
[4] Mechanistic DNA Damage Simulations in Geant4-DNA Part 1: A parameter study in a simplified geometry, N. Lampe et al., Phys. Med. 48 (2018) 135-145 - link
[5] Critical review of rate constants for reactions of hydrated electrons, hydrogen atoms and hydroxyl radicals (·OH/·O-) − in aqueous solution, G. V. Buxton et al., J. Phys. Chem. Ref. Data. 17 (1988) 513–886 - link